Fluorophore: Key tools in molecular imaging
Fluorophores are indispensable in molecular biology, enabling fluorescent labelling. Such fluorescent chemical groups can be attached to large biomolecules like DNA, RNA, proteins and more. This fluorescent labelling technique allows fluorescence-based imaging of biological processes in research and diagnostics. While the term fluorophore refers broadly to any light-emitting molecule, fluorochromes specifically describe dye-based fluorescent molecules commonly used in biological and biochemistrial applications. These fluorophores are widely used in e.g fluorescence microscopy, flow cytometry, high throughput screening, Real-Time and qPCR, sequencing machines and many more molecular DNA and RNA labelling techniques.
What is a fluorophore?
Definition and function
A fluorophore is a molecule capable of absorbing light at a specific wavelength and then emitting it at a longer wavelength. This fluorescent property makes them essential in biological imaging, allowing researchers to track cellular processes, detect biomolecules and improve diagnostic accuracy. Thereby, fluorescent chemical groups are attached to large biomolecules by bioconjugation technologies such as click chemistry. This process is called fluorescent labelling. The fluorophore serves then as a very sensitive marker for molecules like DNA, RNA, proteins etc.. These fluorophores can be visualized via imaging technologies such as fluorescence microscopy, flow cytometry, high throughput screening, Real-Time and qPCR machines or NGS sequencing machines.
Fluorophore vs. fluorochrome
While all fluorochromes are fluorophores, not all fluorophores are fluorochromes. Fluorochromes typically refer to synthetic dye-based fluorophores that are commonly used in biochemical staining techniques. These dyes are usually attached to DNAs or RNAs and visualized using laboratory equipment such as fluorescence microscopy and flow cytometry. In contrast, fluorophores also include naturally occurring fluorescent proteins (e.g. eGFP, mScarlet, Fluc) and advanced materials such as quantum dots.
Applications of fluorophores in molecular biology
Fluorescence microscopy
Fluorophores are widely used in fluorescence microscopy to label and visualize specific cellular structures, proteins, and organelles. They enable both live-cell imaging and fixed-sample analysis, providing insights into cellular dynamics and biological interactions.
A typical example to analyze fixed-samples are EdU cell proliferation assays. The technology is based on the labeling of DNA with EdU (5-ethynyl-2′-deoxyuridine), a nucleoside analog that is incorporated into the replicating DNA of living cells and its conjugation to a fluorochrome in situ.
Figure: Fluorescence microscopy imaging of replicating HeLa cells detected with the EdU Cell Proliferation Kit (BCK-EdU488IM100). Green signal: EdU labelling with 6-FAM (FITC alternative) a fluorophore emitting at 516 nm; blue signal: DNA counterstaining with DAPI or 4′,6-diamidino-2-phenylindole, a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA.
Observing biological processes in real time, i.e. live cell imaging, requires planning and careful handling of living cells during the experiment. To avoid phototoxicity and to minimize the wavelength range, live cell imaging should be performed with a sensitive detector (ideally a cooled CCD camera) and qualified fluorochromes. In addition, a constant temperature in the system and a constant volume of solution in the imaging vessel will help to avoid focus drift. Lentiviral particles encoding fluorescent proteins, cell-permeable fluorochromes or fluorescent nanoparticles can be used, for example, to detect apoptosis, analyze cell viability and health, monitor hypoxia, track reactive oxygen species, monitor the function of calcium indicators and culture nerve and stem cells.
Flow cytometry
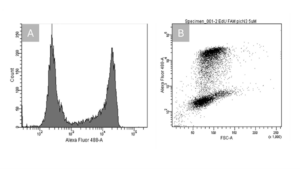
Flow cytometry is a widely used technique to analyze cells pass through a laser. Whereby fluorochromes label cell surface markers or intracellular labelling, allowing for rapid identification and sorting of cell populations based on their fluorescence characteristics. E.g. you can analyze cell viability of e.g. HeLa cells. Therefore, cells have been incubated with the nucleoside analog EdU (5-ethynyl-2’-deoxyuridine), which incorporates into newly synthesized DNA. A click chemistry reaction is then performed, labeling the EdU with the fluorophore e.g. 6-FAM, a fluorophore analog of the Alexa Fluor 488.
Figure: A.) The first peak between 102 and 103 represents cells without EdU incorporation, indicating a lack of DNA synthesis (cell death). These cells exhibit low fluorescence because they did not incorporate the fluorophore 6-FAM. The second peak, shifted to the right (between 104 and 105), represents cells that have actively synthesized DNA and incorporated EdU, showing higher fluorescence due to the fluorophore 6-FAM labeling. B.) In the scatter plot, two distinct cell populations are visible: proliferating, larger, and more complex cells at the top, and non-proliferating, smaller, and less complex cells at the bottom. The clear separation confirms the successful distinction between cells that are undergoing DNA synthesis (and thus incorporated EdU and fluorophore 6-FAM labeling) and those that are not.
Fluorescence in situ hybridization
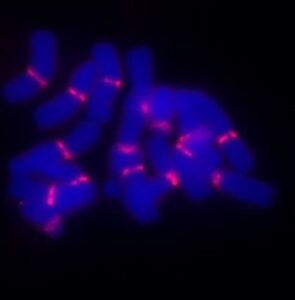
Fluorophores are used to detect specific DNA or RNA sequences within tissues, aiding in diagnostics and genetic research. Fluorophores are essential in fluorescence in situ hybridization (FISH), where they are conjugated to DNA or RNA probes (complementary sequence to the genes of interest) to detect specific genetic sequences within cells or tissues. This application is essential for genetic diagnostics, oncology, and chromosomal research.
Figure: Left: Mitotic rye chromosomes after pre-hybridization click with the fluorophore 5-TAMRA azide labelled centromere repeat FISH probe Bilby (582 bp, in red). Right: FISH-experiments from Arabidopsis-type telomere probes labelled with the fluorophore 5-TAMRA azide.
Protein and nucleic acid labeling
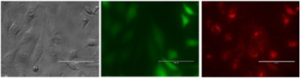
For protein detection unnatural amino acid derivatives, such as 4-Azido-L-phenylalanine, is used e.g. for genetic code expansion in protein biosynthesis as it can be incorporated into the proteins. Due to the azide-moiety, this molecule allows for specific bioconjugation of various functionalities such as e.g. PEGs, antibodies or fluorochromes, by click chemistry or Staudinger ligation. Furthermore, biotin is a useful label for protein detection because of its extraordinarily strong binding to streptavidin. Biotinylated probes are generally used to tag antibodies, antigens or peptides for subsequent detection using enzyme-conjugated streptavidin. Fluorophore labelled streptavidin-biotin detection is a standard technique used e.g. in sandwich ELISA assays. However, biotin is also used to label DNA or RNA like fluorophores. Nucleic acid labelling is becoming an increasingly important research tool. In the last 10 years in particular, RNA labelling has become an interesting tool because it can be used to analyze biodistribution at the cellular level as well as in whole organisms. In experimental setups mRNAs labelled with fluorochromes and coding or fluorescent proteins are used to test biodistribution and protein translation. Click chemistry can be used to produce mRNA labelled with fluorophores specifically at the 5′-end, 3′-end or within the ORF. In the figure below, HeLa cells were transfected with eGFP-encoding mRNA labelled with a Cy3 fluorochrome only at the poly(A) tail. The study shows intact mRNA in the cells, high translation efficiency of the labelled mRNA and possible biodistribution analysis.
Figure: Microscopy images of HeLa cells transfected with eGFP mRNA labeled with Cy3 fluorophore in the poly(A) tail. Left to right: bright field, green fluorescence, and Cy5 channel. Scale bar: 100 mm
Types of fluorophores
Organic dyes
Dye-based fluorochromes, such as fluorescein and rhodamine, are small synthetic molecules commonly used in staining applications due to their bright emission and tunable properties.
Fluorescent proteins
Naturally occurring fluorophores like fluorescent protein (e.g. eGFP, mScarlet, Fluc) and its derivatives are widely used for live-cell imaging and molecular tracking.
Quantum dots
Quantum dots are nanocrystal-based fluorophores known for their exceptional brightness, broad absorption spectra, and resistance to photobleaching, making them ideal for long-term imaging applications.
Selection criteria for fluorophores and fluorochromes
Choosing the right fluorophore depends on several factors:
Excitation and Emission Wavelengths:
The fluorophore of choice should match the available filter sets and laser sources in instruments.
Photostability: Fluorophores with high resistance to photobleaching are perfect for extended imaging experiments.
Multiplexing: When using multiple fluorophores, spectral overlap (emission) of the fluorophores should be minimized to prevent signal interference.
Quantum Yield & Brightness: Higher quantum yield ensures stronger fluorescence signals for enhanced sensitivity.
Environmental Sensitivity: Some fluorophores are pH-sensitive or prone to quenching in certain conditions, which must be considered.
Challenges in using fluorophores
Photobleaching
Dark storage of fluorophore is important because prolonged exposure to excitation light can degrade fluorophore signals over time. Strategies such as dark brown glass storage vials, anti-fade reagents and optimized imaging settings help mitigate photobleaching.
Background signal
Biological samples may exhibit natural auto fluorescence such as mitochondria, potentially interfering with fluorescent markers (fluorophores). Careful fluorophore selection and appropriate controls help distinguish true signals from background noise. Also switch on dyes such as 3- Azido-7-hydroxycoumarin can help to avoid background to noise ratio.
Innovations and future directions of fluorophores
Near-Infrared (NIR) fluorophores are advanced fluorophores which enable deep-tissue imaging. The main advantage is that they have minimal background noise, making them valuable in medical diagnostics and in vivo imaging.
Super-resolution microscopy dyes (fluorophores) are designed for special machines like Stochastic Optical Reconstruction Microscopy (STORM) and Photoactivation Localization Microscopy (PALM). They allow researchers to visualize cellular structures at nanometer resolution.
Smart fluorophores are environment-sensitive fluorophores that change emission based on pH, ion concentration, or molecular interactions are expanding the capabilities of fluorescence imaging.
Switch-on fluorophores such as 3- Azido-7-hydroxycoumarin has the special property to become fluorescent only when it is click ligated. The formaton of a triazole ring expands the electron system of the fluorophores and enables fluorescence. Therefore, there is no background signal of the click unligated leftover fluorophores.