
Sensitive EdU Cell Proliferation Assay for Flow Cytometry
ClickTech Sensitive EdU Cell Proliferation Kit for Flow Cytometry
| Size | Catalog No. | Price |
|---|---|---|
| Dye 488 / 50 Assays | BCK-EdUPro488FC50 | € 525,00 |
| Dye 647 / 50 Assays | BCK-EdUPro647FC50 | € 525,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
2-8 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Excitation (max)
Dye 488: 496 nm | Dye 647: 643 nm
-
Emission (max)
Dye 488: 516 nm | Dye 647: 662 nm
-
Ɛ (max)
Dye 488: 83.000 cm-1M-1 | Dye 647: 250.000 cm-1M-1
-
Preparation/Handling
please see user manual of the kit
Product Information
High‑Resolution Detection of DNA Synthesis with Minimal Cytotoxicity
ClickTech Sensitive EdU Cell Proliferation Kits for Flow Cytometry quantifies DNA replication at the single‑cell level using EdU (5‑ethynyl‑2′‑deoxyuridine) incorporation during S phase and bioorthogonal click chemistry for rapid readout. The kit is optimized for bright signal, low background, and cell health, enabling precise proliferation measurements across primary cells and cell lines—even under low growth or stress conditions.
Dual‑color flexibility for multicolor panels
6‑FAM‑picolyl azide (green): Ex 488 nm / Em 516 nm (FITC channel)
Eterneon‑Red 645‑picolyl azide (red): Ex 647 nm / Em 662 nm (APC channel)
This pairing drops cleanly into standard flow panels, leaving ample room for surface markers, intracellular targets, viability dyes, and DNA‑content stains (e.g., PI or 7‑AAD) to combine proliferation with phenotype and cell‑cycle state.
Recommended workflow (flow‑ready)
- EdU pulse: Add EdU during the intended window (e.g., 15–120 min). Short pulses capture S‑phase entry; longer pulses provide cumulative labeling.
- Fix & permeabilize: Standard aldehyde fixation followed by mild permeabilization retains antigenicity for multicolor panels.
- Click detection: Incubate with the picolyl‑azide dye cocktail (choose green or red). Wash and proceed to surface or intracellular staining as required.
- Acquire & analyze: Gate singlets → exclude dead cells → quantify EdU‑positive events; overlay with DNA‑content dyes (PI/7‑AAD) or cell‑cycle models if desired.
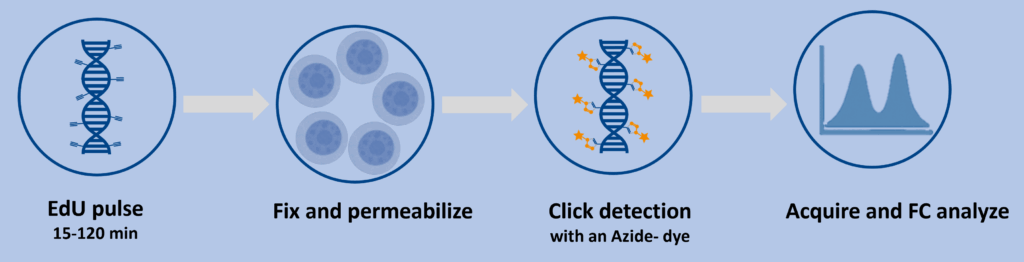
Controls that save time: No‑EdU control, dye‑only control, unstained, and single‑color controls for compensation. For cell‑cycle overlays, include a DNA‑content‑only control.
What makes this kit different
Exceptional sensitivity: Optimized labeling chemistry and picolyl‑azide dyes produce high signal‑to‑noise even in rare or slow‑cycling populations.
Minimal cytotoxicity: The click step is formulated to use substantially less copper than conventional EdU protocols, helping maintain viability and preserving epitopes for downstream staining.
Fast, antibody‑free detection: Fixation and gentle permeabilization give the dye access to DNA without acid, heat, or DNase, so surface and intracellular epitopes remain intact for multicolor immunostaining.
Built for flow cytometry: Clear positive/negative separation, compatibility with standard 488 nm and 640–647 nm lasers, and smooth integration into high‑throughput plate formats.
Reproducible single‑tube workflow: A straightforward pulse‑label → fix/perm → click‑detect sequence typically completes in about two hours, supporting screens and time‑course studies.
Applications
- Cell proliferation kinetics & S‑phase fraction (primary cells and lines)
- Immunology: T‑cell activation/proliferation, memory subset dynamics
- Drug discovery/toxicology: compound response, cytostatic/cytotoxic effects
- Genotoxicity & DNA damage research (combine with DNA‑content dyes)
- Pulse‑chase experiments to follow cell‑cycle progression over time
Performance & compatibility notes
Multicolor panels: Position EdU in FITC (6 FAM) or APC (Eterneon Red 645) to keep PE based channels free for phenotyping.
DNA content dyes: Compatible with PI or 7 AAD for G0/G1, S, and G2/M staging alongside EdU incorporation.
Primary cells: Reduced copper exposure supports more delicate samples and extended acquisition times.
LITERATURE
Analysis of Cell Proliferation and Homeostasis Using EdU Labeling. 2020, Methods Mol Biol.
https://doi.org/10.1007/978-1-4939-2809-5_18
BONCAT-FACS-Seq reveals the active fraction of a biocrust community undergoing a wet-up event. Front. Microbiol. 2023.
https://doi.org/10.3389/fmicb.2023.1176751
Efficient Tandem Copper‐Catalyzed Click Synthesis of Multisugar‐Modified Oligonucleotides. Angewandte Chemie International Edition, e202405161.
https://doi.org/10.1002/anie.202405161
FAQ
-
Does this method require copper?
Yes, the reaction is copper‑catalyzed, but the kit chemistry is tuned for lower copper levels than conventional protocols, which helps maintain cell health.
-
Can I combine this with surface and intracellular immunostaining?
Yes. Mild fixation/permeabilization preserves epitopes, so multicolor immunophenotyping can be run in the same tube.
-
How does this differ from BrdU assays?
It does not require acid/heat denaturation or anti‑BrdU antibodies. That reduces processing time and helps preserve antigenicity, improving multiplex compatibility.
-
Which dye should I choose?
Use 6‑FAM when your panel is tight on red channels, and Eterneon‑Red 645 when FITC needs to remain free. Both deliver high sensitivity; selection is typically panel‑driven.
-
What pulse time should I start with?
A common starting point is ~10 µM EdU for ~2 h, then optimize upward or downward for your specific cell type and biology.

