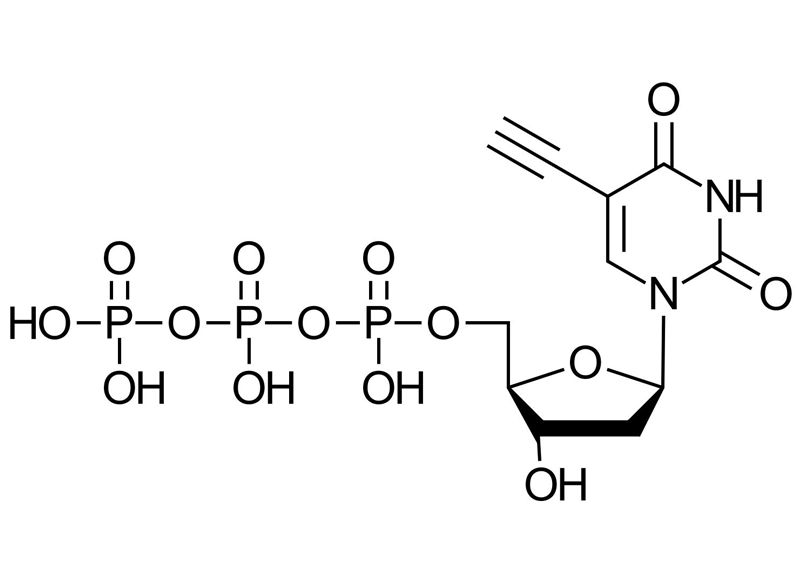
5-Ethynyl-dUTP (5-EdUTP)
Modified triphosphate for incorporation in PCR reaction
| Size | Catalog No. | Price |
|---|---|---|
| 1 µmol | BCT-08-S | € 120,00 |
| 5 µmol | BCT-08-L | € 380,00 |
Chemical Properties
-
Molecular Formula
C11H15N2O14P3
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C
-
Molecular Weight
492.16 g/mol
-
Purity
≥ 95% (HPLC)
-
Physical State
100 mM clear colorless solution in water (pH 7.5)
-
CAS Number
111289-87-3
-
Additional name
EdUTP; 5-Ethynyl-2’-deoxyuridine-5’-triphosphate
-
Absorption (max)
λmax = 288 nm
-
Ɛ (max)
12,000 cm-1M-1
Product Information
A Chemically Modified Triphosphate for DNA Labeling via Click Chemistry
5-Ethynyl-dUTP is a modified deoxyuridine triphosphate featuring an ethynyl group at the 5-position of the uracil base. It enables site-specific DNA labeling through Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) and is compatible with enzymatic DNA synthesis using thermostable polymerases.
The molecular handle for DNA Labeling and Biotechnology
5-Ethynyl-dUTP (5-EdUTP) is a nucleotide triphosphate analog designed for enzymatic incorporation into DNA. The ethynyl group at the C5 position of the uracil base introduces a chemically orthogonal handle that allows for bioorthogonal conjugation via CuAAC. Once incorporated into DNA by DNA polymerases, the alkyne moiety can be selectively reacted with azide-functionalized probes, enabling precise post-synthetic labeling.
This reagent is widely used in molecular biology, cell biology, diagnostics, and sequencing workflows where controlled DNA modification is required.
Scientific Challenges in DNA Labeling
Modified nucleotide triphosphates such as 5-Ethynyl-dUTP offer powerful tools for DNA labeling, but their use in enzymatic reactions like PCR presents several scientific and technical challenges:
Polymerase Selectivity
DNA polymerases are optimized for natural nucleotides. Even small modifications at the nucleobase can affect recognition and incorporation efficiency. The success of modified dNTPs depends heavily on the polymerase’s tolerance for structural variation.
Steric and Electronic Effects
Modifications at the nucleobase (e.g., ethynyl groups at C5 of uracil) must be positioned to avoid interference with base pairing and enzyme-substrate interactions. Bulky or charged groups can hinder incorporation or reduce fidelity.
Enzyme-Specific Compatibility
Not all polymerases incorporate modified dNTPs equally. Family A polymerases (e.g., Taq) and Family B polymerases (e.g., Pwo, Deep Vent, KOD) differ in their active site architecture. Empirical testing is often required to identify suitable enzymes for each modification.
Reduced Amplification Efficiency
Incorporation of modified dNTPs may slow down elongation or reduce overall yield. Optimization of reaction conditions, including Mg²⁺ concentration, extension time, and dNTP ratios, is often necessary.
Template and Primer Design Considerations
Modified dNTPs may affect hybridization dynamics. Primers and templates must be designed to ensure efficient initiation and elongation in the presence of modified nucleotides.
Downstream Compatibility
DNA containing modified bases must be validated for compatibility with downstream applications such as sequencing, cloning, or hybridization. Some modifications may interfere with enzyme-based detection or ligation.
Advantages of 5-Ethynyl-dUTP
Bioorthogonal Reactivity: The ethynyl group enables selective conjugation with azide-functionalized probes via CuAAC under mild, aqueous conditions.
Polymerase Compatibility: Efficiently incorporated by thermostable DNA polymerases such as Taq and Pwo, allowing use in PCR, primer extension, and cell-free synthesis.
Versatile Utility: Applicable in fixed and live-cell assays, flow cytometry, fluorescence microscopy, and NGS workflows.
Applications in Research and Diagnostics
- Molecular Biology & Biotechnology: Direct incorporation into DNA during enzymatic synthesis and compatible with PCR, qPCR, and DNA polymerase-based workflows
- NGS & Library Preparation: Introduces clickable handles for downstream conjugation and enrichment
- Drug Screening & Toxicology: Quantifies DNA synthesis in response to compounds or treatments
- Biosensor Development: Enables site-specific attachment of reporter molecules or capture tags to DNA
- dNTP Pool Quantification: Used in click-based assays to measure endogenous dTTP levels without radioisotopes
LITERATURE
Synthesis of Highly Modified DNA by a Combination of PCR with Alkyne-Bearing Triphosphates and Click Chemistry, J. Gierlich et al., 2007, Chem. Eur. J., Vol. 13(34), p. 9486–9494.
https://doi.org/10.1002/chem.200700502
Directed DNA Metallization, G. A. Burley et al., 2006, J. Am. Chem. Soc., Vol. 128(5), p. 1398–1399.
https://doi.org/10.1021/ja055517v
Identification and characterization of nucleobase-modified aptamers by click-SELEX, F. Pfeiffer et al., 2018, Nature Protocols, Vol. 13(5), p. 1153–1180.
https://doi.org/10.1038/nprot.2018.023
Quantitation of deoxynucleoside triphosphates by click reactions, C. Y. Huang et al., 2020, Scientific Reports, Vol. 10(1), p. 611.
https://doi.org/10.1038/s41598-020-57463-3
FAQ
-
Which polymerases are compatible with EdUTP?
Taq and Pwo polymerases show high incorporation efficiency.
-
Can I use 5-EdUTP in NGS workflows?
Yes. It introduces clickable handles for downstream functionalization and enrichment.

